About HybridKla
Lysine lactylation (Kla), a novel lactate-derived post-translational modification (PTM), is involved in a myriad of biological processes and complex diseases.
Here, we report a friendly online service.We manually collected 23,984 Kla sites across 7,297 proteins from 14 species to construct the comprehensive Kla benchmark dataset.
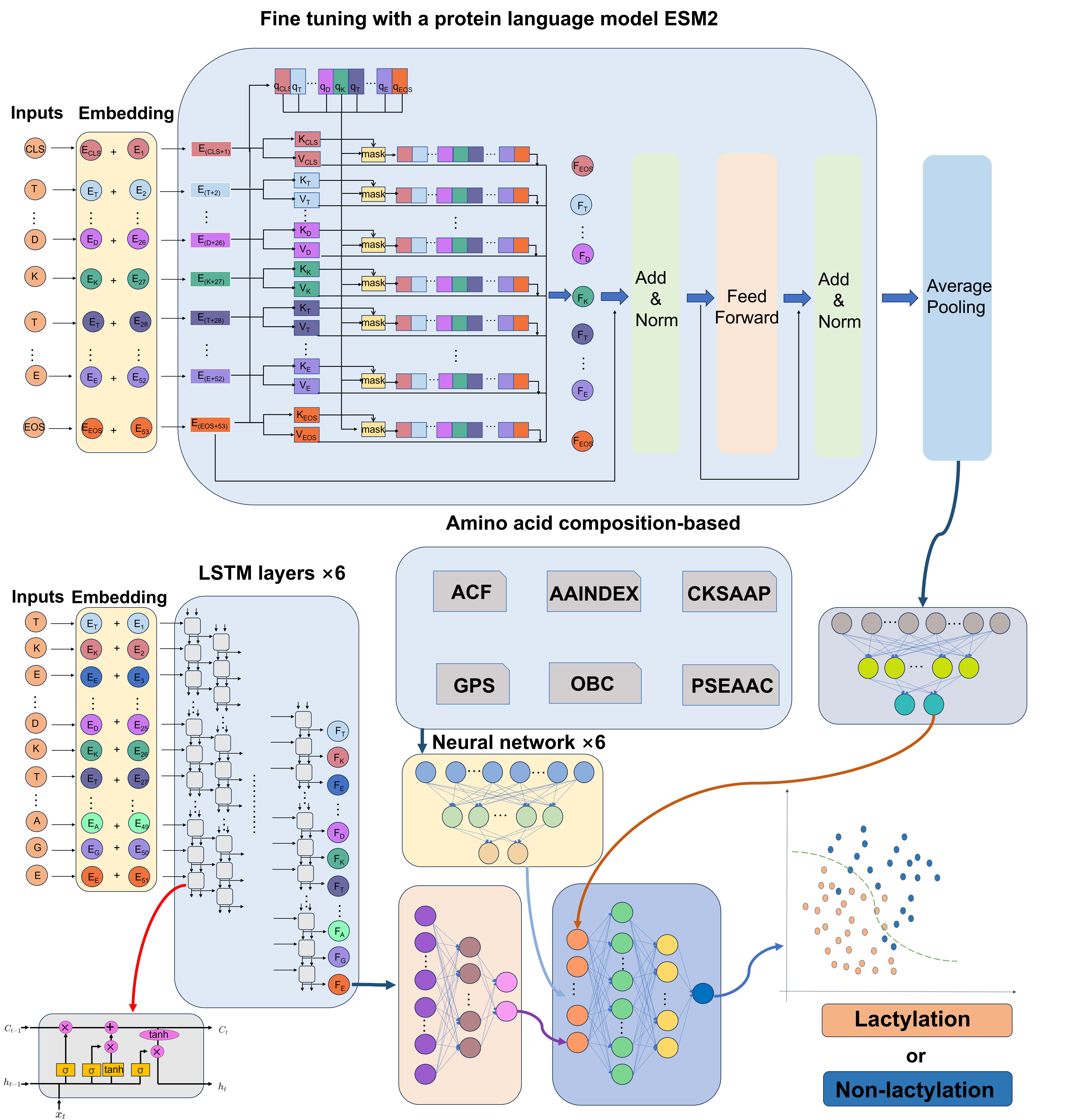
We designed a multi-feature hybrid system that synergistically combines two automated encoding strategies—derived from a fine-tuned protein language model ESM2 and a six-layer long short-term memory (LSTM) module—with a composition-based framework incorporating six handcrafted descriptors: ACF, AAINDEX, CKSAAP, GPS, OBC, and PSEAAC. Leveraging this hybrid encoding system together with deep learning, we developed a novel prediction tool named HybridKla for precise identification of Kla sites, achieving an area under the curve (AUC) value of 0.8460, which represents a >28.90% improvement of the AUC value compared to existing tools (0.8460 versus 0.6563).
For users, one or multiple protein sequences could be inputted in the FASTA format, and the output will be shown in a list. For the help of online service and the tutorial, please refer to the User Guide page.